Perturbs points with a uniform perturbation in a circle.
Note that r can either be one distance, or a distance per data point.
See also
Other point pertubation:
mask_grid(),
mask_voronoi(),
mask_weighted_random()
Examples
x <- cbind(
x = c(2.5, 3.5, 7.2, 1.5),
y = c(6.2, 3.8, 4.4, 2.1)
)
# plotting is only useful from small datasets!
# grid masking
x_g <- mask_grid(x, r=1, plot=TRUE)
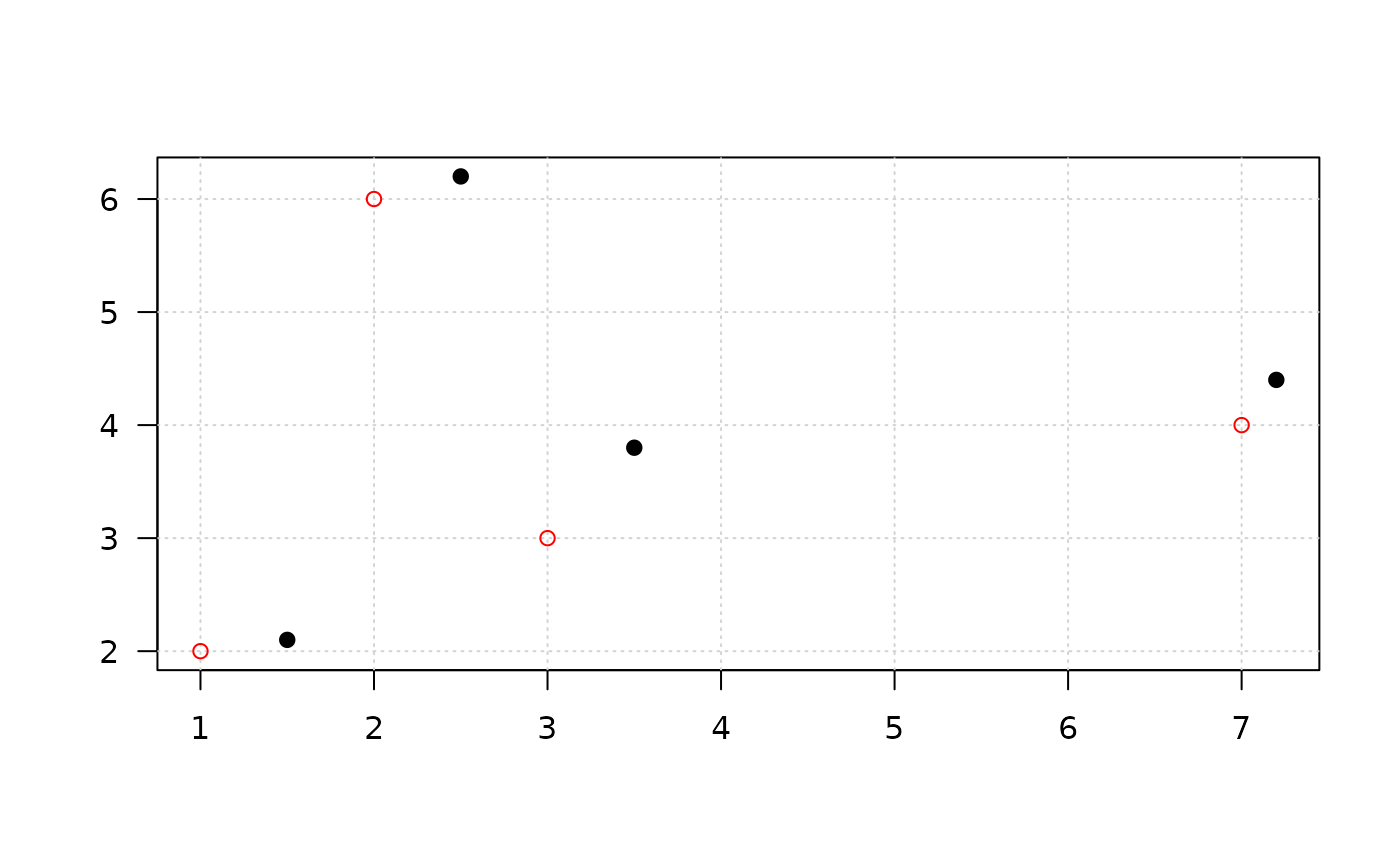 # random pertubation
set.seed(3)
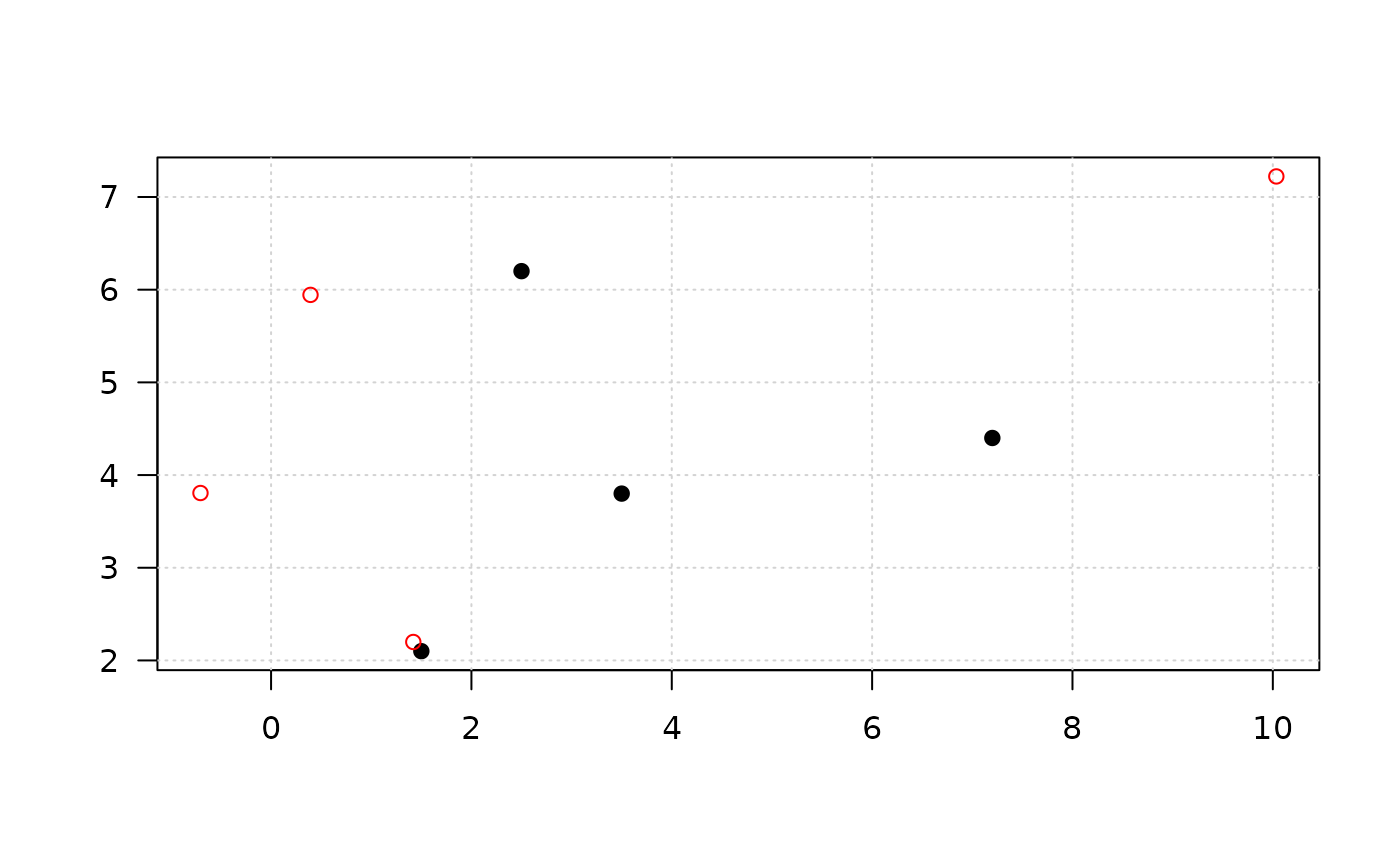
x_r <- mask_random(x, r=1, plot=TRUE)
# random pertubation
set.seed(3)
x_r <- mask_random(x, r=1, plot=TRUE)
 x_d <- mask_donut(x, r_min=1, r_max=2, plot=TRUE)
x_d <- mask_donut(x, r_min=1, r_max=2, plot=TRUE)
 # \donttest{
if (requireNamespace("FNN", quietly = TRUE)){
# weighted random pertubation
x_wr <- mask_weighted_random(x, k = 2, r = 4, plot=TRUE)
}
# \donttest{
if (requireNamespace("FNN", quietly = TRUE)){
# weighted random pertubation
x_wr <- mask_weighted_random(x, k = 2, r = 4, plot=TRUE)
}
 if ( requireNamespace("FNN", quietly = TRUE)
&& requireNamespace("sf", quietly = TRUE)
){
# voronoi masking, plotting needs package `sf`
x_vor <- mask_voronoi(x, r = 1, plot=TRUE)
}
if ( requireNamespace("FNN", quietly = TRUE)
&& requireNamespace("sf", quietly = TRUE)
){
# voronoi masking, plotting needs package `sf`
x_vor <- mask_voronoi(x, r = 1, plot=TRUE)
}
 # }
# }