cond_barplot() conditions all variables on x by quantile binning and
shows the median or mean of the other variables for each x.
cond_barplot(
data,
x = NULL,
n = 100,
min_bin_size = NULL,
overlap = NULL,
ncols = NULL,
fill = "#2f4f4f",
auto_fill = FALSE,
show_bins = FALSE,
type = c("median", "mean"),
...
)Arguments
- data
a
data.frameto be binned- x
charactervariable name used for the quantile binning- n
integernumber of quantile bins.- min_bin_size
integerminimum number of rows/data points that should be in a quantile bin. If NULL it is initiallysqrt(nrow(data))- overlap
logicalifTRUEthe quantile bins will overlap. Default value will beFALSE.- ncols
The number of column to be used in the layout.
- fill
The color to use for the bars.
- auto_fill
If
TRUE, use a different color for each category- show_bins
If
TRUE, show the bins on the x-axis.- type
The type of statistic to use for the bars.
- ...
Additional arguments to pass to the plot functions
Value
A list of ggplot objects.
See also
Other conditional quantile plotting functions:
cond_boxplot(),
cond_heatmap(),
funq_plot()
Examples
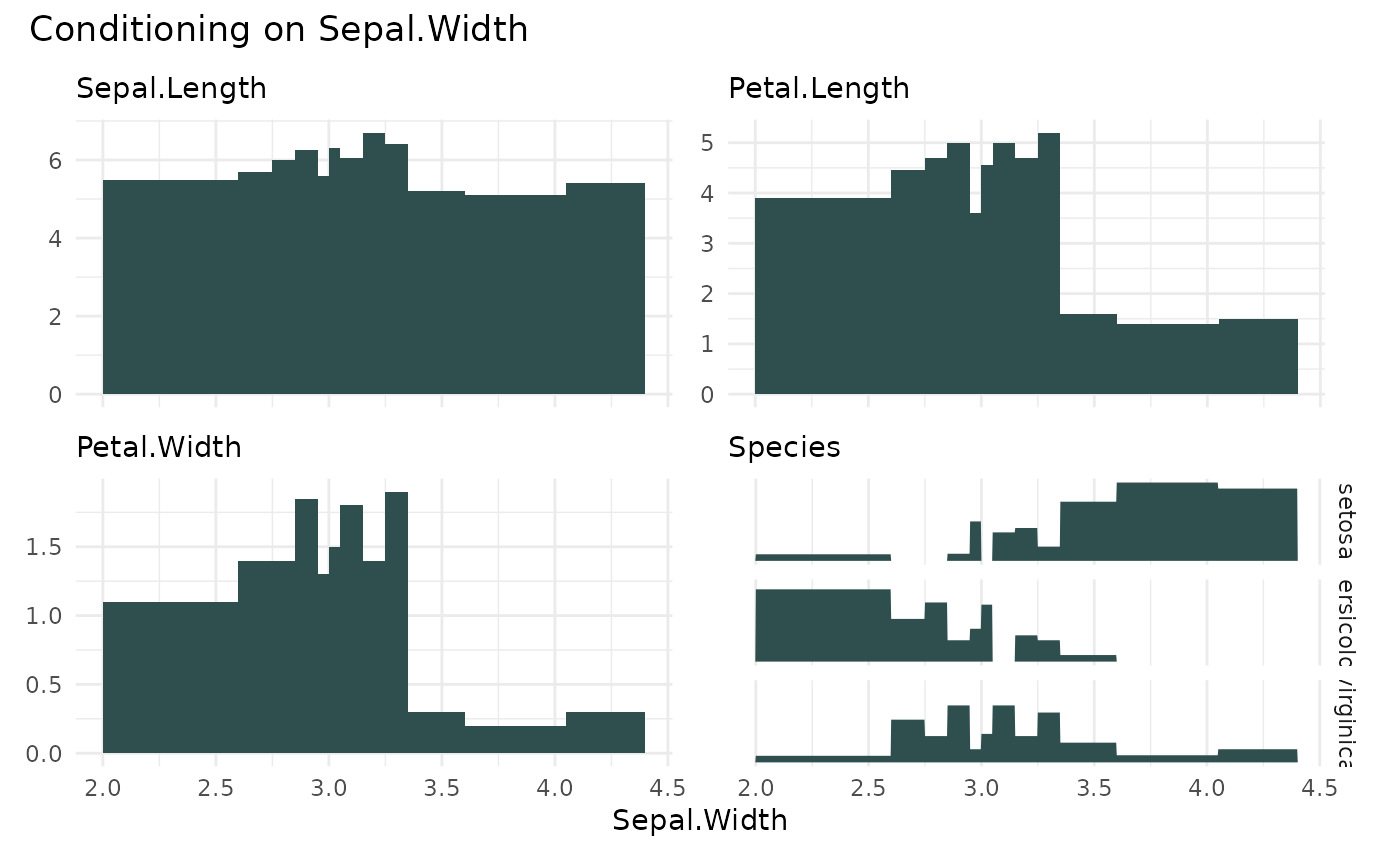
# plots the expected median conditional on Sepal.Width
cond_barplot(iris, "Sepal.Width", n = 12)
 # \donttest{
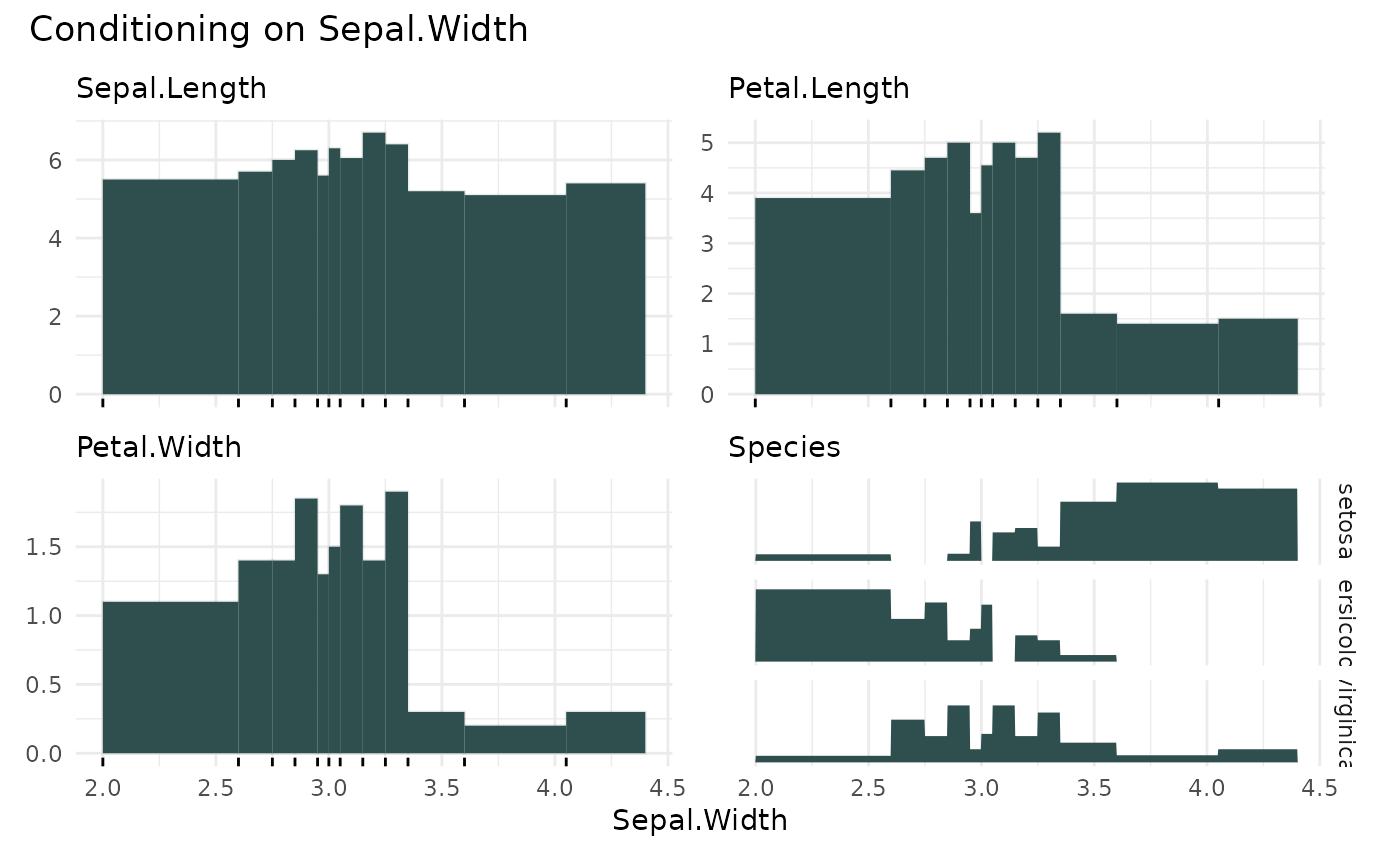
# plots the expected median
cond_barplot(iris, "Sepal.Width", n = 12, show_bins = TRUE)
# \donttest{
# plots the expected median
cond_barplot(iris, "Sepal.Width", n = 12, show_bins = TRUE)
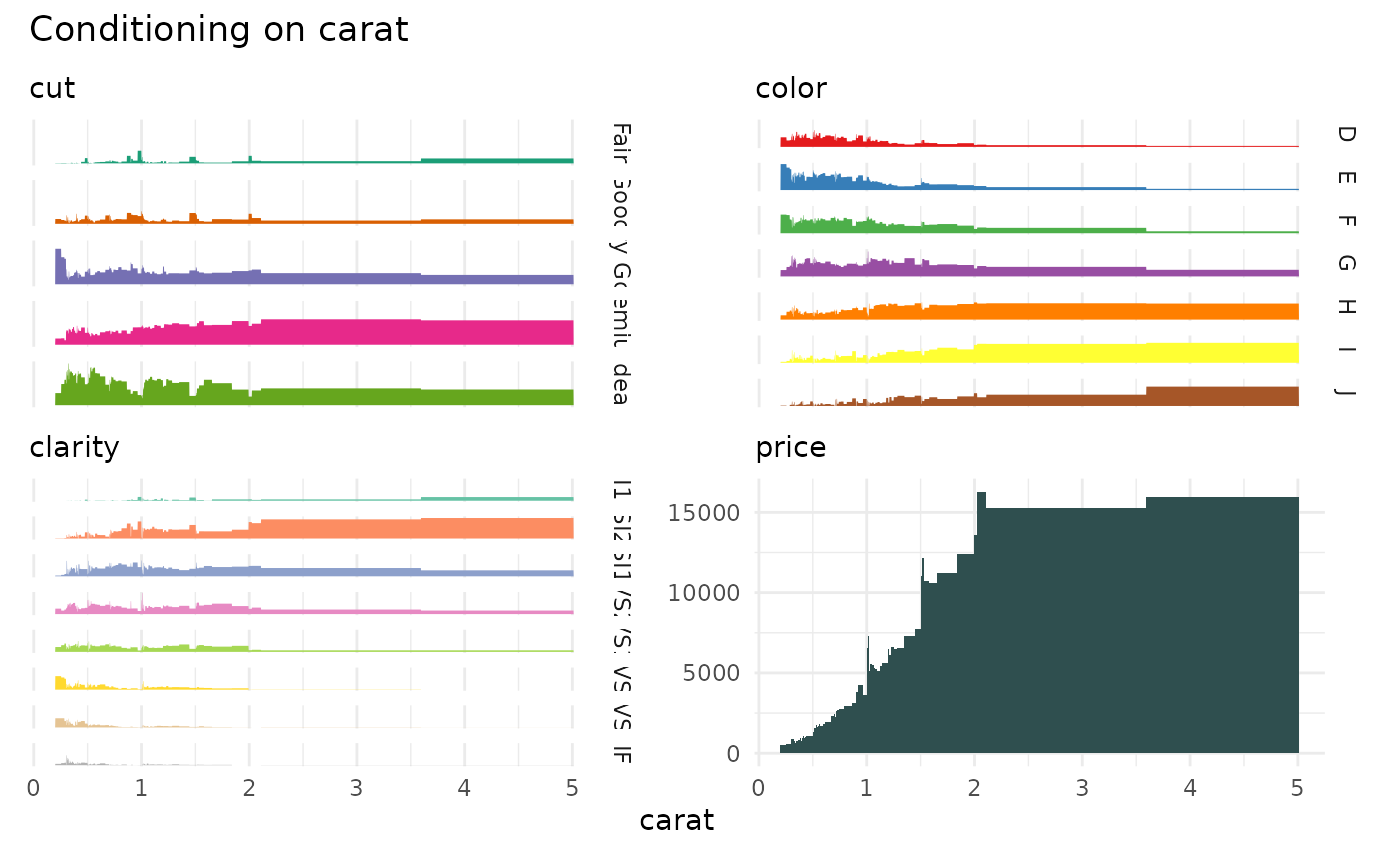
 data("diamonds", package="ggplot2")
cond_barplot(diamonds[c(1:4, 7)], "carat", auto_fill = TRUE)
data("diamonds", package="ggplot2")
cond_barplot(diamonds[c(1:4, 7)], "carat", auto_fill = TRUE)
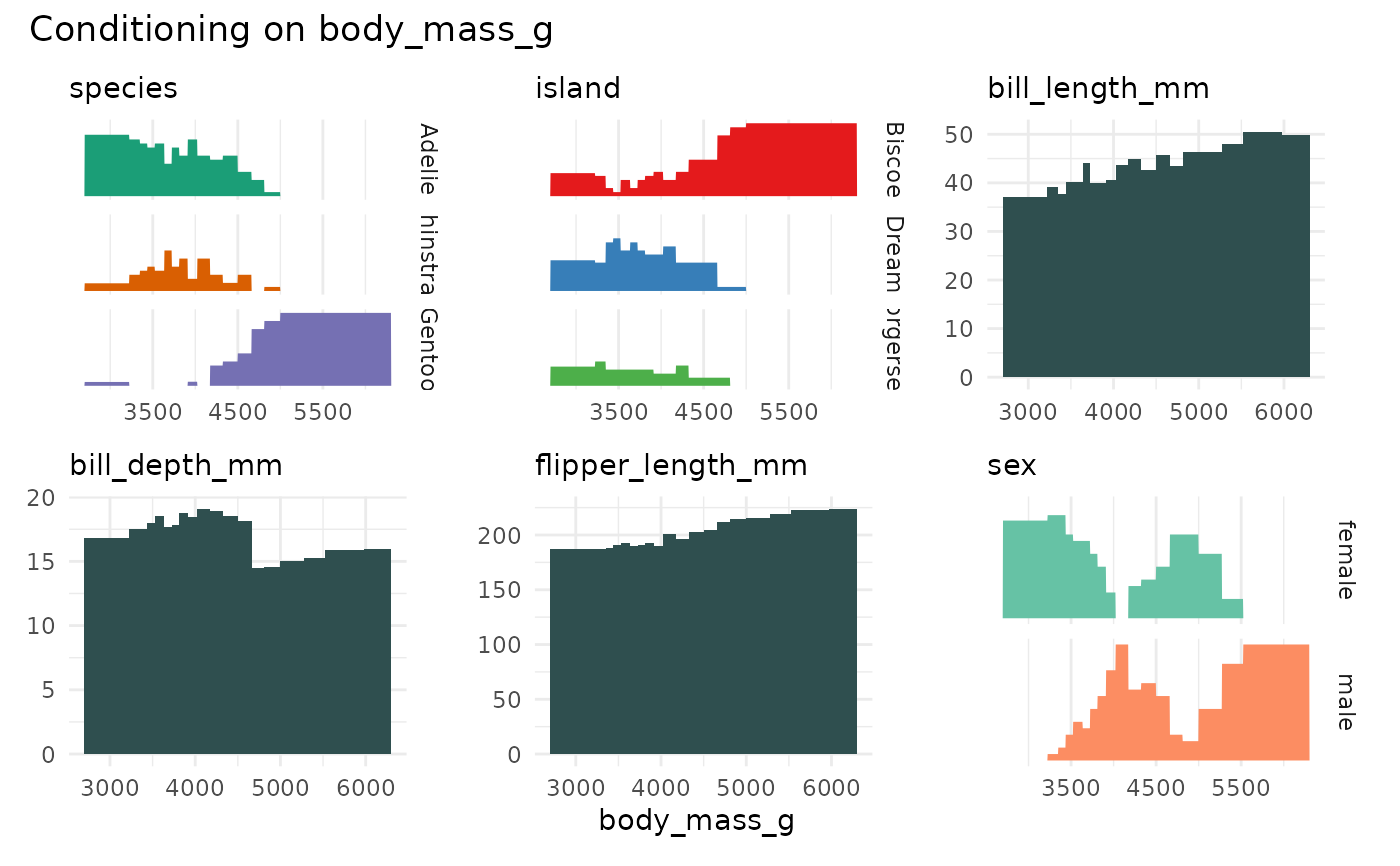
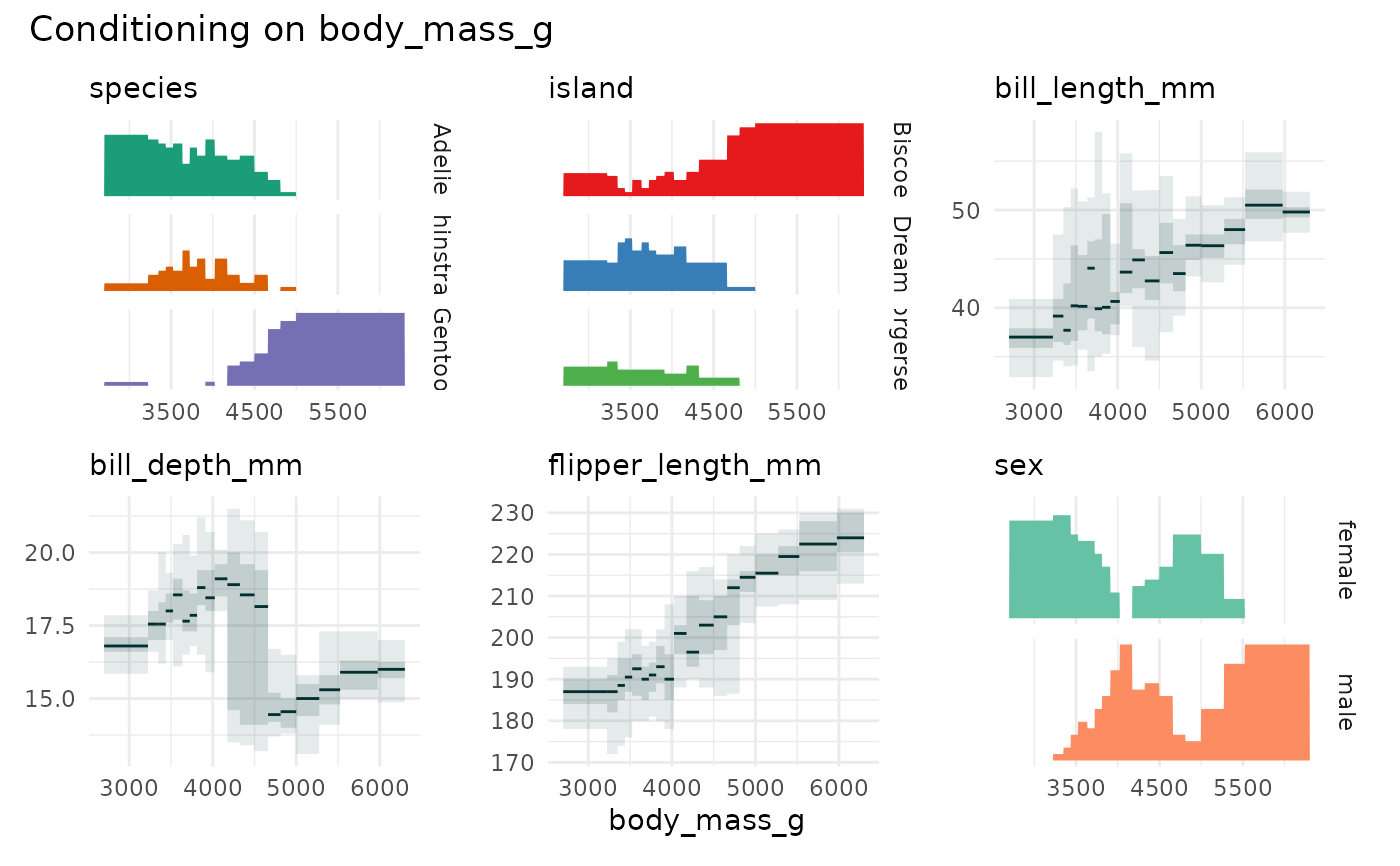
 if (require(palmerpenguins)) {
p <- cond_barplot(penguins[1:7], "body_mass_g", auto_fill = TRUE)
print(p)
# compare with qbin_boxplot
p <- cond_boxplot(penguins[1:7], "body_mass_g", auto_fill = TRUE)
print(p)
}
#> Loading required package: palmerpenguins
#> `overlap` not specified, using `overlap=FALSE`
#> `min_bin_size`=18, using `n=19`
if (require(palmerpenguins)) {
p <- cond_barplot(penguins[1:7], "body_mass_g", auto_fill = TRUE)
print(p)
# compare with qbin_boxplot
p <- cond_boxplot(penguins[1:7], "body_mass_g", auto_fill = TRUE)
print(p)
}
#> Loading required package: palmerpenguins
#> `overlap` not specified, using `overlap=FALSE`
#> `min_bin_size`=18, using `n=19`
 #> `overlap` not specified, using `overlap=FALSE`
#> `min_bin_size`=18, using `n=19`
#> `overlap` not specified, using `overlap=FALSE`
#> `min_bin_size`=18, using `n=19`
 # }
# }